Chapter 7 Designs for improving precision: blocking with single experimental unit
7.1 Introduction
In our discussion of the vendor example in Section 3.2, we observed huge gains in power and precision when allocating each vendor to one of two samples per mouse, and comparing the resulting enzyme levels for each pair of samples and then averaging these differences.
This design is an example of blocking, where we organize the experimental units into groups (or blocks), such that units within the same group are more similar than those in different groups. For \(k\) treatments, a block size of \(k\) experimental units per group allows us to randomly allocate each treatment to one unit per block and we can estimate any treatment contrast within each block and then average these estimates over the blocks. Even though large differences might exist between blocks, between-block variation is is removed by this analysis strategy if they affect all treatments in the block equally. This reduces the residual variance for contrasts and tests, resulting in increases precision and power without increases in sample size. As R. A. Fisher noted:
Uniformity is only requisite between objects whose response is to be contrasted (that is, objects treated differently). (Fisher 1971, 33)
To be effective, blocking requires that we find some property by which we can group our experimental units such that variances within each group are smaller than between groups. This property can be intrinsic, such as animal body weight, litter, or sex, or non-specific to the experimental units, such as day of measurement in a longer experiment, batch of some necessary chemical, or the machine used for incubating samples or recording measurements (Sect. 7.2.4).
Our main design is the randomized complete block design (RCBD), where each block has as many units as there are treatments, and we randomly assign each treatment to one unit in each block (Sect. 7.2). We can extend the design to a generalized randomized complete block design (GRCBD) when we use more than one replicate of each treatment per block (Sect. 7.2.6.1). If the block size is smaller than the number of treatments, a balanced incomplete block design (BIBD), still allows treatment allocations balanced over blocks such that all pair contrasts are estimated with the same precision (Sect. 7.3).
Several blocking factors can be combined in a design by nesting—allowing estimation of each blocking factor’s contribution to variance reduction—or crossing—allowing simultaneous removal of several independent sources of variation. We postpone discussion of two classic designs with crossed blocks—latin squares and Youden squares—to the next Chapter ??.
7.2 The randomized complete block design
7.2.1 Experiment and data
We continue with our example of how three drug treatments in combination with two diets affect enzyme levels in mice. To keep things simple, we only consider the low fat diet for the moment, so the treatment structure only contains Drug with three levels. Our aim is to improve the precision of contrast estimates and increase the power of the omnibus \(F\)-test for Drug. For this, we need to identify a blocking factor such that (i) we can determine the groups of mice before we start randomizing the drug treatment and (ii) the within-group variance is much lower than the between-groups variance. A common choice for blocking mice is by litter, since sibling mice often show more similar responses as compared to mice from different litters (Perrin 2014). Litter sizes are typically in the range of \(5-7\) in mice (Watt 1934), which would easily allow us to select groups of three mice from each litter for our experiment.
Our experiment is illustrated in Figure 7.1(A): we use \(b=8\) litters of \(n=3\) mice each, resulting in an experiment size of \(N=bn=24\) mice. In contrast to a CRD, our randomization is restricted since we require that each treatment occurs exactly once per litter, and we randomize drugs independently for each litter.
The data are shown in Figure 7.1(B), where we connect the three observed enzyme levels in each block by a line, akin to an interaction plot. The vertical dispersion of the lines indicates that enzyme levels within each litter are systematically different from those in other litters. The lines are roughly parallel, which shows that all three drug treatments are affected equally by these systematic differences, there is no litter-by-drug interaction, and treatment contrasts are unaffected by systematic 4differences between litters.
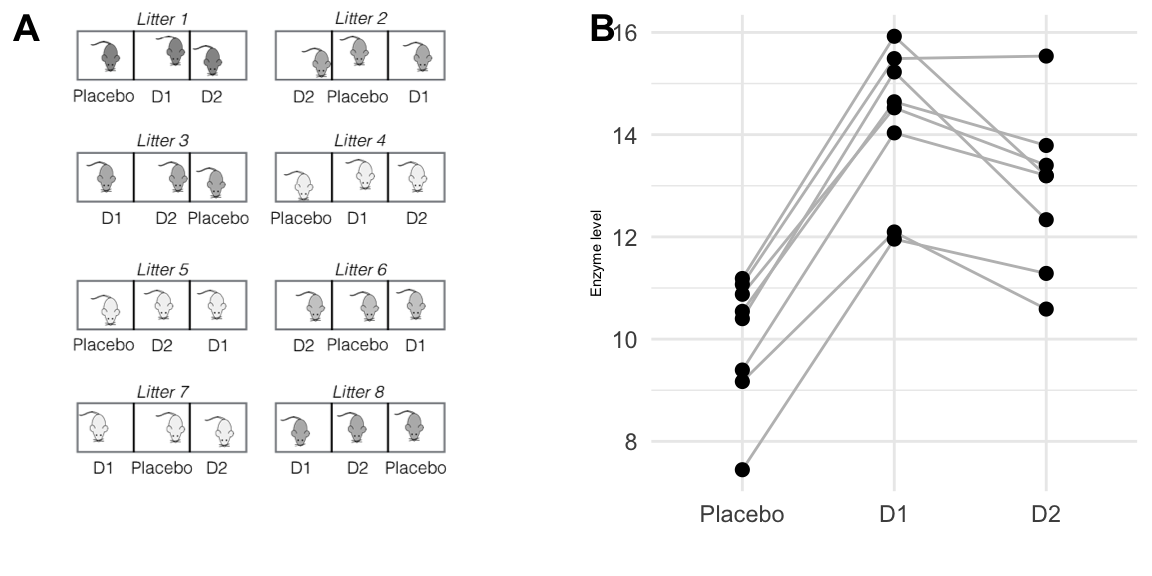
Figure 7.1: Comparing enzyme levels for placebo, D1, and D2 under low fat diet using randomized complete block design. A: data layout with eight litters of size three, treatments independently randomized in each block. B: observed enzyme levels for each drug; lines connect responses for mice from the same litter.
7.2.2 Model and Hasse diagram
Hasse diagram
Deriving the experiment structure diagram in Figure 7.2 poses no new challenges: the treatment structure contains the single treatment factor Drug with three levels while the unit structure contains the blocking factor (Litter), which groups the experimental units from (Mouse). We consider the specific eight litters in our experiment as a random sample, which makes both unit factors random. The factors Drug and (Litter) are crossed and their combinations are levels of the interaction factor (Litter:Drug); since (Litter) is random, so is the interaction. The randomization allocates levels of Drug on (Mouse), and the experiment structure is similar to a two-way factorial design, but (Litter) originates from the unit structure, is random, and is not allocated to (Mouse), but rather groups levels of (Mouse) by an intrinsic property of each mouse.
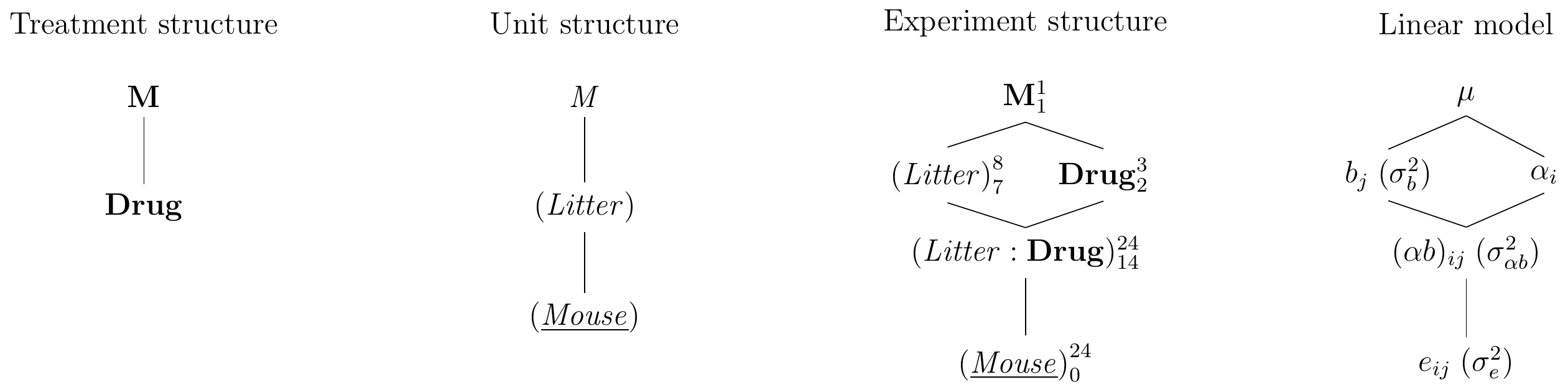
Figure 7.2: Randomized complete block design for determining effect of two different drugs and placebo using eight mice per drug and a single response measurement per mouse. Each drug occurs once per block and assignment to mice is randomized independently for each block.
The block-by-treatment interaction
Each treatment occurs only once per block, and the variations due to interactions and residuals are completely confounded. If we want to analyse data from this design, we need to assume that the block-by-treatment interaction is negligible. We can then merge the interaction and residual factors and use the sum of their variation for estimating the residual variance. The design is then called a randomized complete block design (RCBD).
An appreciable block-by-treatment interaction means that the differences in enzyme levels from litter to litter depend on the drug treatment. Treatment contrasts are then litter-specific and this systematic heterogeneity of treatment effects complicates the analysis and precludes a straightforward interpretation of the results. Such interaction is likely caused by other biologically relevant factors that influence the effect of (some) treatments but that have not been accounted for in our experiment and analysis.
Since we cannot test the interaction factor, we require a non-statistical argument to justify ignoring the interaction. Note that we have full control over which property to use for blocking and—in contrast to a two-way ANOVA—are not interested in contrasts on the blocking factor. In many cases, subject-matter knowledge allows us to exclude interactions between our chosen blocking factor and the treatment factor. In our particular case, for example, it seems unlikely that the litter effect affects drugs differently, which justifies treating the litter-by-drug interaction as negligible; our design is then an RCBD.
Linear model
Because the blocking factor levels are random, so are the cell means \(\mu_{ij}\). Moreover, there is a single observation per cell and each cell mean is confounded with the residual for that cell; a two-factor cell means model is therefore not useful for an RCBD.
The full parametrized model (including block-by-treatment interaction) is \[ y_{ij} = \mu + \alpha_i + b_j + (\alpha b)_{ij} + e_{ij}\;, \] where \(\mu\) is the grand mean, \(\alpha_i\) the deviation of the average response for drug \(i\) compared to the grand mean, and \(b_j\) is the random effect of block \(j\). We assume that the residuals and block effects are normally distributed as \(e_{ij}\sim N(0,\sigma_e^2)\) and \(b_j\sim N(0,\sigma_b^2)\), and are all mutually independent. The interaction of drug and block effects is a random effect, too, with distribution \((\alpha b)_{ij}\sim N(0,\sigma^2_{\alpha b})\); the model is shown in Figure 7.2 (right).
For an RCBD, the interaction is completely confounded with the residuals; its parameters \((\alpha b)_{ij}\) cannot be estimated from the experiment without replicating treatments within each block. If interactions are negligible, \((\alpha b)_{ij}=0\) and we arrive at the additive model \[\begin{equation} y_{ij} = \mu + \alpha_i + b_j + e_{ij} \tag{7.1}\;. \end{equation}\] The average response to treatment level \(i\) is then \(\mu_i=\mu+\alpha_i\); it is unbiasedly estimated as \[ \hat{\mu}_i = \frac{1}{b}\sum_{j=1}^b y_{ij}\;, \] and this estimator has variance \(\text{Var}(\hat{\mu}_i)=(\sigma_b^2+\sigma_e^2)/b\). We then base our contrast analysis on these estimated cell means, effectively using a cell means model for the treatment factor after correcting for the block effects.
Importantly, contrasts between treatment group means are estimated much more precisely than the group means themselves. For example, the average difference between placebo and \(D1\) is \(\mu_1-\mu_2=(\mu+\alpha_1)-(\mu+\alpha_2)\), which we estimate as \[\begin{align*} \widehat{\mu_1-\mu_2} &= \frac{1}{b}\sum_{j=1}^b \left(y_{1j}-y_{2j}\right)\\ &= \frac{1}{b}\sum_{j=1}^b\left((\mu+\alpha_1+b_j+e_{1j})-(\mu+\alpha_2+b_j+e_{2j})\right) \\ &= \alpha_1-\alpha_2 + \frac{1}{b}\sum_{j=1}^b (e_{1j}-e_{2j})\;. \end{align*}\] Since the random block effects \(b_j\) and the residuals \(e_{ij}\) are all independent, this estimator is unbiased. Its variance is \[ \text{Var}\left(\widehat{\mu_1-\mu_2}\right) = \frac{1}{b}\sum_{j=1}^b\left(y_{1j}-y_{2j}\right)\;, = 2\sigma_e^2/b\;, \] and the between-block variance \(\sigma_b^2\) is completely removed from this estimate because of the covariance between observations in the same block.
Analysis of variance
The total sum of squares for the RCBD with an additive model decompose into sums of squares for each factor involved, such that \[ \text{SS}_\text{tot} = \text{SS}_\text{drug} + \text{SS}_\text{block} + \text{SS}_\text{res}\;, \] and the degrees of freedom decompose accordingly. Mean squares are formed by dividing each sum of squares term by its corresponding degrees of freedom.
The treatment factor is tested by comparing its mean squares to those of the residuals using the \(F\)-statistic \[ F = \frac{\text{SS}_\text{drug}/\text{df}_\text{drug}}{\text{SS}_\text{res}/\text{df}_\text{res}}\;. \] For \(k\) treatment factor levels and \(b\) blocks, this test statistic has a \(F_{k-1,\, (b-1)(k-1)}\) distribution under the null-hypothesis \(H_0:\mu_1=\mu_2=\cdots =\mu_k\) that all treatment group means are equal.
We derive the model specification for R’s aov() function from the Hasse diagrams in Figure 7.2: since fixed and random factors are exclusive to the treatment respectively unit structure, the corresponding terms are 1+drug and Error(litter/mouse); these terms are equivalent to drug and Error(litter) and the model is fully specified as y~drug+Error(litter). The analysis of variance table then contains two error strata: one for the block effect and one for the within-block residuals. We randomized the treatment on the mice within litters, and the block effect stratum therefore contains no treatment effects; all comparisons of drug levels are conducted in the within-block error stratum. The analysis yields
##
## Error: litter
## Df Sum Sq Mean Sq F value Pr(>F)
## Residuals 7 37.08 5.297
##
## Error: Within
## Df Sum Sq Mean Sq F value Pr(>F)
## drug 2 74.78 37.39 84.55 1.53e-08 ***
## Residuals 14 6.19 0.44
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The omnibus \(F\)-test for the treatment factor provides clear evidence that the drugs affect the enzyme levels differently and the differences in average enzyme levels between drugs is about 85 times larger than the residual variance.
The effect size \(\eta^2_\text{drug}=\) 63% is large, but measures the variation explained by the drug effects compared to the overall variation, including the variation removed by blocking. It is more meaningful to compare the drug effect to the within-block residuals alone and ignore the litter variation by using the partial effect size \(\eta_\text{p, drug}^2=\) 92%. This confirms that the overwhelming fraction of the variation of enzyme levels in each litter is due to the drugs acting differently. In other words, the drugs have an appreciable effect that should be easy to detect in contrast analysis, and blocking by litter removed a substantial amount of variation, making comparisons between drugs very precise.
In contrast to a two-way ANOVA with factorial treatment structure, we cannot simplify the analysis to a one-way ANOVA with a single treatment factor with \(b\cdot k\) levels. This is because the blocking factor is random, and the resulting one-way factor would also be a random factor. The omnibus \(F\)-test for this factor is difficult to interpret, and a contrast analysis would be a futile exercise, since we would compare randomly sampled factor levels among each other.
Linear mixed model
In contrast to our previous designs, the analysis of an RCBD requires two variance components in our model: the residual variance \(\sigma_e^2\) and the between-blocks variance \(\sigma_b^2\). The classical analysis of variance handles these by using one error strata per variance component, but this makes subsequent analyses more tedious. For example, estimates of the two variance components are of direct interest, but while the estimate \(\hat{\sigma}_e^2=\text{MS}_{\text{res}}\) is directly available calculated, an estimate for the between-blocks variance is \[ \hat{\sigma}_b^2 = \frac{\text{MS}_{\text{block}}-\text{MS}_{\text{res}}}{n}\;, \] and requires manual calculation. For our example, we find \(\hat{\sigma}_e^2=\) 0.44 and \(\hat{\sigma}_b^2=\) 1.62 based on the ANOVA mean squares.
A more modern alternative is the linear mixed model, which explicitly considers the differenct random factors for estimating variance components and parameters of the linear model in Equation (7.1). Linear mixed models offer a very general and powerful extension to linear regression and analysis of variance, but their general theory and estimation are beyond the scope of this book. For our purposes, we only need a small fraction of their possibilities and we use the lmer() function from package lme4 for all our calculations.9 In specifying a linear mixed model, we use terms of the form (1|X) to introduce a random offset for each level of the factor X; this construct replaces the Error()-term from aov(). The fixed effect part of the model specification remains unaltered. For our example, the model specification is then y~drug+(1|litter), which asks for a fixed effect for each level of Drug, and allows a random offset for each litter. This yields
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: y ~ drug + (1 | litter)
## Data: ddrug
##
## REML criterion at convergence: 66.1
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -1.23932 -0.67405 -0.04512 0.52952 1.67537
##
## Random effects:
## Groups Name Variance Std.Dev.
## litter (Intercept) 1.6181 1.272
## Residual 0.4422 0.665
## Number of obs: 24, groups: litter, 8
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 10.0109 0.5075 9.4020 19.726 5.73e-09 ***
## drugD1 4.2259 0.3325 14.0000 12.709 4.46e-09 ***
## drugD2 2.9057 0.3325 14.0000 8.739 4.83e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) drugD1
## drugD1 -0.328
## drugD2 -0.328 0.500Only the Random effects part is of direct interest for us and provides the two variance components \(\hat{\sigma}_b^2=\) 1.62 (litter) and \(\hat{\sigma}_e^2=\) 0.44 (Residual), in close agreement with the ANOVA results. The Fixed effects part gives the parameter estimates for \(\alpha_i\), but they are of secondary interest and depend on the coding (cf. Section 4.5.2). Note that the degrees of freedom for estimating the intercept (which corresponds to the average enzyme level in the placebo group) is no longer an integer. This is because block effects and residuals are used with different weights in its estimation.
We calculate our familiar ANOVA table from an estimated linear mixed model using anova(lmdrug, ddf="Kenward-Roger") which yields
| Sum Sq | Mean Sq | NumDF | DenDF | F value | Pr(>F) | |
|---|---|---|---|---|---|---|
| drug | 74.78 | 37.39 | 2 | 14 | 84.55 | 1.528e-08 |
For a balanced design, this method gives the same results as the ‘traditional’ analysis of variance. An advantage of linear mixed models, however, is their ability to directly cope with unbalanced data. Colloquially, we can say that the analysis of variance approach provides a convenient and powerful framework to phrase design questions, while the linear mixed model provides a more general and powerful tool for the subsequent analysis.
7.2.3 Contrasts
The emmeans() function readily handles both aov() estimates with error strata and lmer() estimates, and provides one cell mean per treatment level (the three \(\hat{\mu}_i\) in our example). We can therefore define and estimate treatment contrasts exactly as before, and briefly exemplify the procedure for our experiment with three contrasts: the comparison of \(D1\) respectively \(D2\) to placebo, and the comparison of the average of \(D1\) and \(D2\) to placebo. The contrasts are
\[
\Psi(\mathbf{w}_1) = \mu_2-\mu_1
\,,\quad
\Psi(\mathbf{w}_2)=\mu_3-\mu_1
\,,\quad\text{and}\quad
\Psi(\mathbf{w}_3)=\frac{\mu_2+\mu_3}{2}-\mu_1\;.
\]
The following code defines and estimates the contrasts based on the linear mixed model:
# Estimate linear mixed model
lmdrug = lmer(y~drug+(1|litter), data=ddrug)
# Estimate group means
em.lmer = emmeans(lmdrug, ~drug)
# Manually define contrasts
psi = list("D1 - Placebo"=c(-1,+1,0),
"D2 - Placebo"=c(-1,0,+1),
"(D1+D2)/2 - Placebo"=c(-1,+1/2,+1/2))
# Estimate contrasts and 95%-CI
ci.lmer = confint(contrast(em.lmer, psi))| contrast | estimate | SE | df | lower.CL | upper.CL |
|---|---|---|---|---|---|
| ANOVA; aov(y~drug+Error(litter)) | |||||
| D1 - Placebo | 4.23 | 0.33 | 14 | 3.51 | 4.94 |
| D2 - Placebo | 2.91 | 0.33 | 14 | 2.19 | 3.62 |
| (D1+D2)/2 - Placebo | 3.57 | 0.29 | 14 | 2.95 | 4.18 |
| Mixed model: lmer(y~drug+(1|litter)) | |||||
| D1 - Placebo | 4.23 | 0.33 | 14 | 3.51 | 4.94 |
| D2 - Placebo | 2.91 | 0.33 | 14 | 2.19 | 3.62 |
| (D1+D2)/2 - Placebo | 3.57 | 0.29 | 14 | 2.95 | 4.18 |
| INCORRECT: aov(y~drug) | |||||
| D1 - Placebo | 4.23 | 0.72 | 21 | 2.73 | 5.72 |
| D2 - Placebo | 2.91 | 0.72 | 21 | 1.41 | 4.40 |
| (D1+D2)/2 - Placebo | 3.57 | 0.62 | 21 | 2.27 | 4.86 |
The contrast estimates for our experiment are shown in Table 7.1 for an analysis based on aov(), the above analysis using lmer(), and an incorrect analysis of variance that ignores the blocking factor and treats the data as coming from a completely randomized design. Ignoring the blocking factor means that the litter-to-litter variation becomes part of the residual variance. The resulting decrease in precision is clearly visible, even if the degrees of freedom increase.
Contrast analysis makes no sense for contrasts involving the blocking factor since its levels are random. To estimate the difference between the average enzyme level of the first and second litter, for example, only has a useful interpretation, if these two arbitraty litters are again used in a replication of the experiment, which is rarely the case.
7.2.4 Evaluating and choosing a blocking factor
Evaluating a blocking factor
Once the data are recorded, we might be interested to quantify how ‘good’ the blocking performed in the experiment. This information would allow us to better predict the expected residual variance for a power analysis of our next experiment and to determine if we should continue using the blocking factor.
One way of evaluating the blocking is by treating the blocking factor as fixed, and specify the ANOVA model as y~drug+litter. The ANOVA table then contains a row for the blocking factor with an associated \(F\)-test (Samuels, Casella, and McCabe 1991). This test only tells us if the between-block variance is significantly different from zero, but does not quantify the advantage of blocking.
A more meaningful alternative is the calculation of appropriate effect sizes, and we can determine the percentage of variation removed from the analysis by blocking using the effect size \(\eta^2_\text{block}\), which evaluates to 31% for our example. In addition, the partial effect \(\eta^2_\text{p, block}=\) 86% shows that the vast majority of non-treatment sum of squares is due to the litter-to-litter variation and blocking by litter was a very successful strategy.
While \(\eta^2_\text{p, block}\) measures the proportion of variation due to blocking based on the as sums of squares. Alternatively, the intraclass correlation coefficient (ICC) \(\text{ICC}=\sigma_b^2 / (\sigma_b^2+\sigma_e^2)\) uses the proportion of variance directly, and we find an ICC of 79% for our example, confirming that the blocking works well. The ICC is also directly related to the relative efficiency of the RCBD compared to a non-blocked CRD, since \[ \text{RE}(\text{CRD},\text{RCBD}) = \frac{\sigma_b^2+\sigma_e^2}{\sigma_e^2} = \frac{1}{1-\text{ICC}}\;, \] and \(\sigma_b^2+\sigma_e^2\) is the residual variance of a corresponding CRD. From our estimates of the variance components, we find a relative efficiency of 4.66; to achieve the same precision as our blocked design with 8 mice per treatment group would require about 37 mice per treatment group for a completely randomized design. We can alternatively find the relative efficiency as \[ \text{RE}(\text{CRD},\text{RCBD}) = \frac{\widehat{\text{MS}}_\text{res, CRD}}{\text{MS}_\text{res, RCBD}}\;, \] where we estimate the residual mean squares of the CRD from the ANOVA table of the RCBD as a weighted average of the block and the residual sums of squares, weighted by the corresponding degrees of freedom: \[ \widehat{\text{MS}}_\text{res, CRD} = \frac{\text{df}_\text{block}\cdot \text{MS}_\text{block}\, +\, (\text{df}_\text{trt}+\text{df}_\text{res})\cdot \text{MS}_\text{res}}{\text{df}_\text{block}+\text{df}_\text{trt}+\text{df}_\text{res}}\;,. \] For our example, \(\widehat{\text{MS}}_\text{res, CRD}=\) 1.92, resulting in a relative efficiency estimate of RE(CRD,RCBD)=4.34. It differs slightly from our previous estimate because it uses an approximation based on the ANOVA results rather than estimates of the variance components based on the linear mixed model.
Choosing a blocking factor
The main purpose of blocking is to decrease the residual variance for improving the power of tests and precision of estimates by grouping the experimental units before the allocation of treatments, such that units within the same block are more similar than units from different blocks. The blocking factor must therefore describe a property of experimental units that is independent of the treatment outcome, which precludes, e.g., grouping as ‘responders’ and ‘non-responders’ based on the measured response. To ensure a meaningful analysis of treatment contrasts, we must also take care that the blocking factor affects all treatment factor levels equally and does not interact with the treatment factor.
Some examples of grouping units for increased precision using intrinsic properties are (i) litters of animals; (ii) parts of a single animal or plant, such as left/right kidney or leafs; (iii) initial weight; (iv) age or co-morbidities; (v) biomarker values, such as blood pressure, state of tumor, or vaccination status. Properties non-specific to the experimental units include (i) batches of chemicals used for the experimental unit; (ii) device used for measurements; or (iii) date in multi-day experiments. While these also increase precision, they are often also necessary due to the logistics of the experiment to account for systematic difference in measurements based on different batches or taken on different days.
The golden rule for designing experiments with blocks is
Block what you can, randomize what you can not.
As always, logistics of the experiment and other practical considerations should be taken into account when deciding upon blocking and randomization. There is no point in designing an overly complicated blocked experiment that becomes too difficult to implement correctly. On the other hand, there is no harm if we use a blocking factor that turns out to have no or minimal effect on the residual variance.

Figure 7.3: Creating a randomized complete block design from a completely randomized design. Left: a CRD randomizes drugs on mice. Center: introducing a blocking factor ‘above’ groups the experimental units. Right: subdividing each experimental units and randomizing the treatments on the lower level creates a new experimental unit factor ‘below’.
We can think about creating a blocked design by starting from a completely randomized design and ‘splitting’ the experimental unit factor into a blocking and a nested (potentially new) unit factor. Two examples are shown in Figure 7.3, starting from the CRD randomly allocating drug treatments on mice. In the first RCBD (Figure 7.3(center)), we create a blocking factor ‘above’ the original experimental unit factor and group mice into litters. This restricts the randomization of Drug to mice within litters. In the second RCBD (Figure 7.3(right)), we subdivide the experimental unit into smaller units by taking multiple samples per mouse. This re-purposes the original experimental unit factor as the blocking factor and introduces a new factor ‘below’, but requires that we now randomize Drug on (Sample) to obtain an RCBD and not pseudo-replication.
7.2.5 Power analysis and sample size
Power analysis for a randomized complete block design with no block-by-treatment interaction is very similar to a completely randomized design, the only difference being that we use the within-block variance \(\sigma_e^2\) as our residual variance. For a main effect with parameters \(\alpha_i\), the noncentrality parameter is thus \(\lambda_R=n\sum_i \alpha_i^2/\sigma_e^2=n\cdot k\cdot f_R^2\), while it is \(\lambda_C=n\sum_i \alpha_i^2/(\sigma_b^2+\sigma_e^2)=n\cdot k \cdot f_C^2\) for the same treatment structure in a completely randomized design. The two effect sizes are different for the same raw effect size, and \(f_C^2\) is smaller than \(f_R^2\) for the same values of \(n\), \(k\), and \(\alpha_i\); this directly reflects the increased precision in an RCBD.
The denominator degrees of freedom are \((k-1)(b-1)=kb-k-(b-1)\) for an RCBD and we loose \(b-1\) degrees of freedom compared to the \(k(b-1)=tb-k\) degrees of freedom for a CRD. The corresponding loss in power is very small and more than compensated by the simultaneous increase in power due to the smaller residual variance unless the blocking is very ineffective and the number of blocks large.
As an example, we consider a single treatment factor with \(k=\) 3 levels, a total sample size of \(N=b\cdot k=\) 30 (\(n=b\) per treatment factor level), and an effect size of \(f_{C,0}^2=\) 0.0625 for the treatment main effect at a significance level of \(\alpha=5\%\). The residual variance is \(\sigma^2=\) 2. Under a completely randomized design, we have a power of 0.2 for detecting the effect. This power increases dramatically to 0.71 when we organize the experimental units into \(b=\) 10 blocks with between-blocks variance \(\sigma_b^2=\) 1.6 and within-block variance \(\sigma_e^2=\) 0.4. The much smaller residual variance increases the standardized effect size to \(f_{R,0}^2=(\sigma_b^2+\sigma_e^2)/\sigma_e^2\cdot f^2_{C,0}=\) 0.3125.
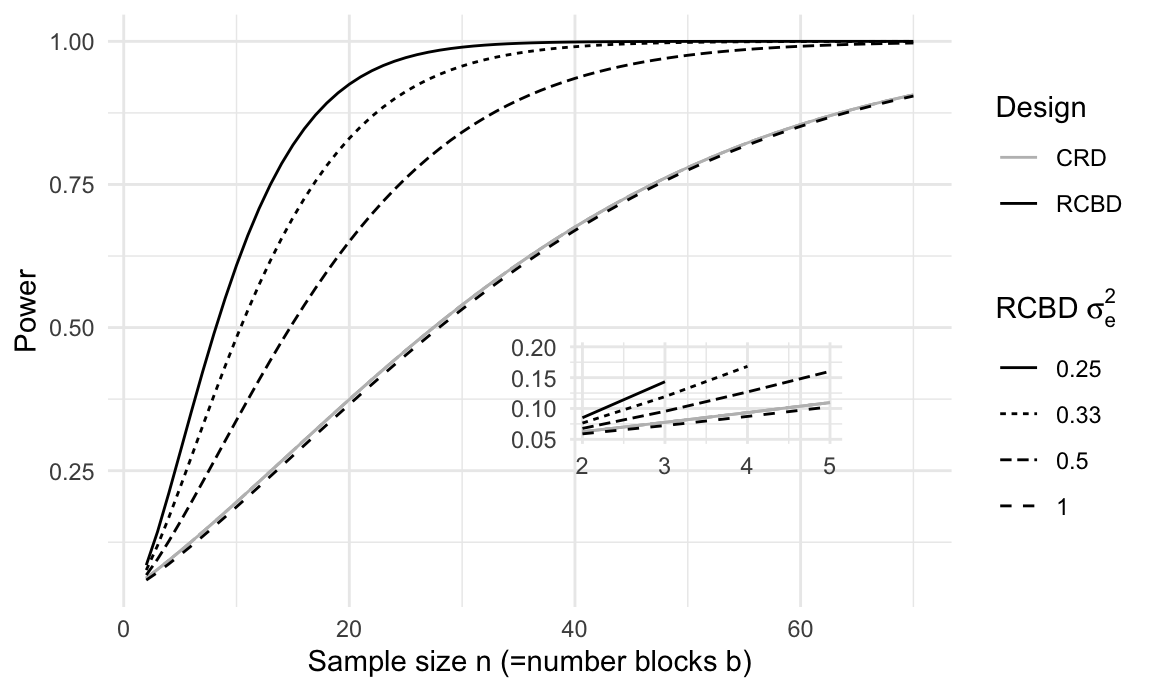
Figure 7.4: Power for different sample sizes n for a completely randomized design with residual variance one and four randomized complete block designs with same overall variance, but four different within-block residual variances. Inset: same curves for small sample sizes show lower power of RCBD than CRD for identical residual variance due to lower residual degrees of freedom in the RCBD.
The relation between power and within-block variance is shown in Figure 7.4 for five scenarios with significance level \(\alpha=5\%\) and an omnibus-\(F\) test for a main effect with \(k=\) 3 treatment levels. The solid grey curve shows the power for a CRD with residual variance \(\sigma_e^2+\sigma_b^2=1\) as a function of sample size per group and a minimal detectable effect size of \(f^2_\text{C}=\) 0.0625. The black curves show the power for an RCBD with four different within-block variances from \(\sigma_e^2=0.25\) to \(\sigma_e^2=1\). For the same sample size, the RCBD then has larger effect size \(f^2_\text{R}\), which translates into higher power. The inset figure shows that an RCBD has negligibly less power than a CRD for the same residual variance of \(\sigma_e^2=1\), due to the loss of \(b-1\) degrees of freedom.
7.2.6 Variations
We briefly discuss two variations of the RCBD which are important in practice. First, we extend the RCBD to allow for multiple replicates of each treatment in each block. Second, we use the standard RCBD idea with a small factorial treatment structure and consider our drug-diet example with two treatment factors, with mice blocked by litter.
7.2.6.1 Replication within blocks
In some scenarios, it is necessary to use more than one replicate of each treatment per block. This is typically the case if our blocking factor is non-specific and introduced to capture grouping due to the logistics of the experiment. For example, we might conduct our drug comparisons in two different laboratories. We then replicate a full CRD in the two laboratories, leading to a design with lab as a blocking factor, but multiple observations of each treatment in each laboratory. This design is helpful if the logistics of implementing the experiment is too laborious or time-consuming for a single laboratory. This might happen if measuring each mouse requires substantial time, and we are concerned that the time difference between first and last mouse measured is so large that it compromises the comparison of the treatment effects. We would expect that the response values show systematic differences between the two labs, due to calibration of measurement devices, implementation of bench protocols, or experience of technical staff. We account for these differences by using the laboratories as levels of a blocking factor, but each laboratory will have several replicates of the same drug treatment.
Another purpose for using a multi-laboratory experiment is to broaden the inference for our experiment. We expect that the training of staff, the kits, buffers, and chemicals used, and the protocols might differ between the labs. If we nevertheless find stable estimates of contrasts between laboratories, then we demonstrated experimentally that our scientific findings are independent of ‘typical’ alterations in the protocol or chemicals used. In fact, replicating pre-clinical experiments in at least two laboratories was recently shown to greatly increase the reproducibility of the results (Karp 2018), and is a standard method for proficiency testing of laboratories.
The resulting design is called a generalized randomized complete block design (GRCBD) and its diagrams are shown in Figure 7.5 for eight mice per drug and two laboratories. Each mouse is used in exactly one laboratory and (Lab) thus groups (Mouse). We use the same drugs in both laboratories, making (Lab) and Drug crossed factors. The replication of treatments in each block allows estimation of the lab-by-drug interaction, which provides information about the stability of the effects and contrasts.
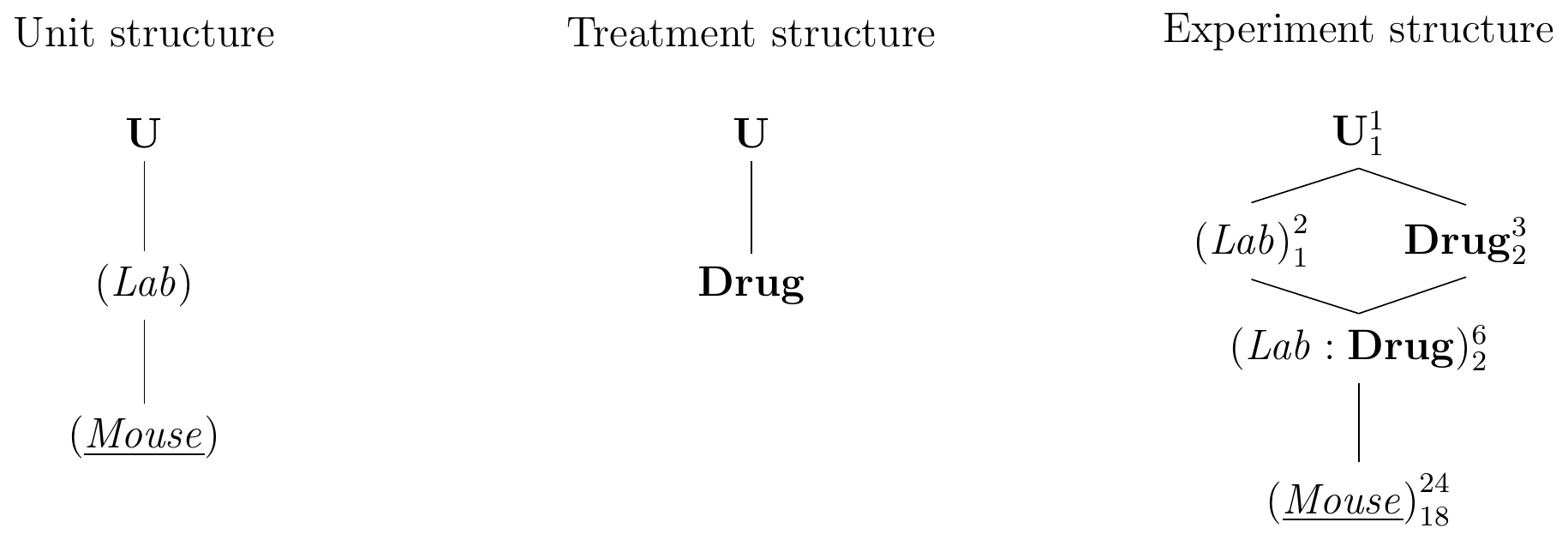
Figure 7.5: A (generalized) randomized complete block design (GRCBD) with four mice per drug and laboratory. Completely randomized design for estimating drug effects of three drugs replicated in two laboratories, with lab-by-drug interaction assumed zero.
The linear model with interaction is
\[
y_{ijk} = \mu + \alpha_i + l_j + (\alpha l)_{ij} + e_{ijk}\;,
\]
where \(\alpha_i\) are the (fixed) treatment effect parameters, \(l_j\) the random block effect parameters with \(l_j\sim N(0,\sigma_l^2)\), \((\alpha l)_{ij}\sim N(0,\sigma_{\alpha l}^2)\) describes the interaction of treatment \(i\) and block \(j\), and \(e_{ijk}\sim N(0,\sigma_e^2)\) are the residuals. The specification for a linear mixed model is y~drug+(1|lab)+(1|lab:drug) for our example.
If the lab-by-treatment interaction is negligible with \(\sigma_{\alpha l}^2\approx 0\), we remove the interaction term from the analysis and arrive at the additive model. Then, (Lab) acts as a blocking factor just like (Litter) in the previous example and eliminates the lab-to-lab variation from treatment contrasts.
If the interaction is appreciable and cannot be neglected, then the contrasts between drugs are different between the two labs, and we cannot determine generalizable drug effects. Now, Lab:Drug is the first random factor below Drug and provides the factor for comparing levels of Drug, which resembles a sub-sampling, where (Mouse) is sub-sampled from Lab:Drug.
7.2.6.2 Factorial treatment structure
It is straightforward to extend an RCBD from a single treatment factor to other factorial treatment structures by crossing the entire treatment structure with the blocking factor. Each block then contains one full replicate of the factorial design, and the required block size rapidly increases with the number of factors and factor levels. In practice, only smaller factorials can typically be blocked by this method since heterogeneity between experimental units often increases with block size, diminishing the advantages of blocking. We discuss more sophisticated designs for blocking factorials that overcome this problem by using only a fraction of all treatment combinations per block in Chapter 9.
We illustrate the extension using our drug-diet example from Chapter 6 where three drugs were combined with two diets. The six treatment combinations can still be accommodated when blocking by litter, and using four litters of size six results in the experimental layout shown in Figure 7.6 with 24 mice in total. Already including a medium-fat diet would require atypical litter sizes of nine mice, however.
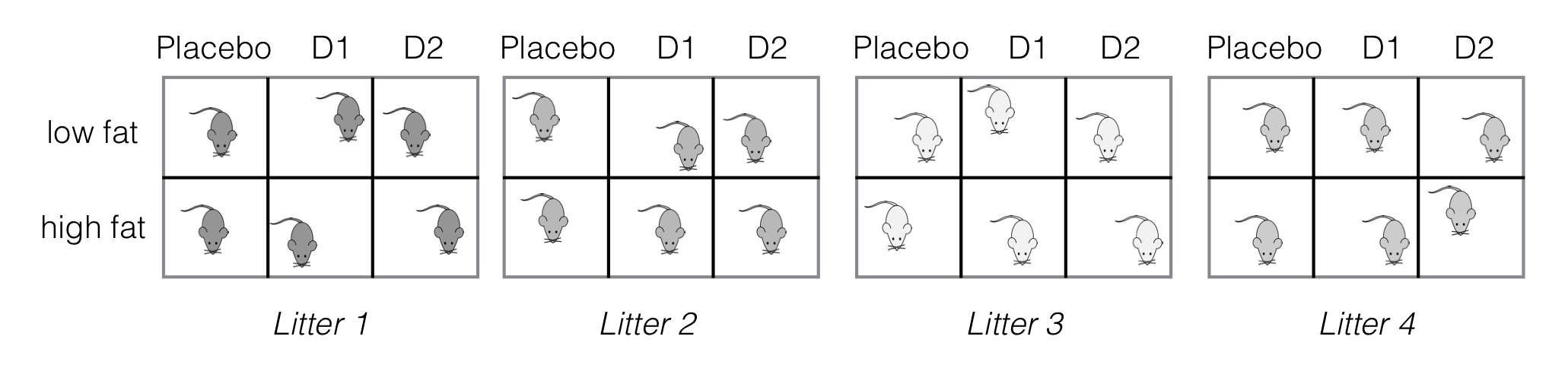
Figure 7.6: Experiment layout for three drugs under two diets, four litter blocks. Completely randomized design with two-way crossed treatment structure within each block.
Model and Hasse diagram
Constructing the Hasse diagrams in Figure 7.7 follows the same procedure as before and provides no difficulties. The blocking factor is crossed with the full treatment structure—both treatment factors and the treatment interaction factor. We assume that our blocking factor does not differentially change the diet or drug effects and consequently ignore all three block-by-treatment interactions (Litter:Diet), (Litter:Drug), and (Litter:Diet:Drug).
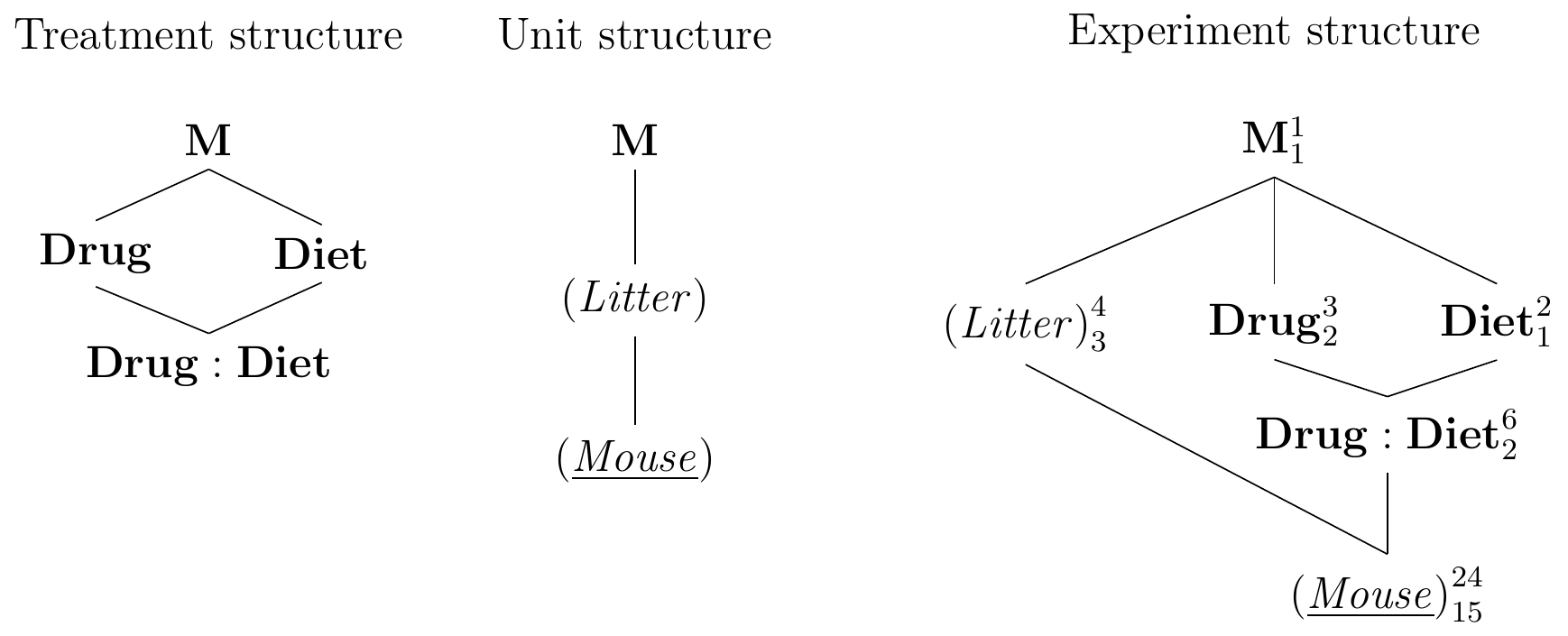
Figure 7.7: Randomized complete block design for determining effect of two different drugs and placebo on two different diets using four mice per drug and a single response measurement per mouse. All treatment combinations occur once per block and are randomized independently.
The corresponding linear model is \[ y_{ijk} = \mu + \alpha_i + \beta_j + (\alpha\beta)_{ij} + b_k + e_{ijk}\;, \] where \(\alpha_i\) and \(\beta_j\) are the main effects of drug and diet, respectively, \((\alpha\beta)_{ij}\) the interaction effects between them, \(b_k\sim N(0,\sigma_b^2)\) is the random block effect, and \(e_{ijk}\sim N(0,\sigma_e^2)\) are the residuals.
The enzyme levels \(y_{ijk}\) are shown in Figure 7.8 as black points in an interaction plot separately for each block. We can clearly see the block effects, which systematically shift enzyme levels for all conditions. After estimating the linear model, the shift \(b_k\) can be estimated for each block, and the grey points show the resulting ‘normalized’ data \(y_{ijk}-\hat{b}_k\) after accounting for the block effects.
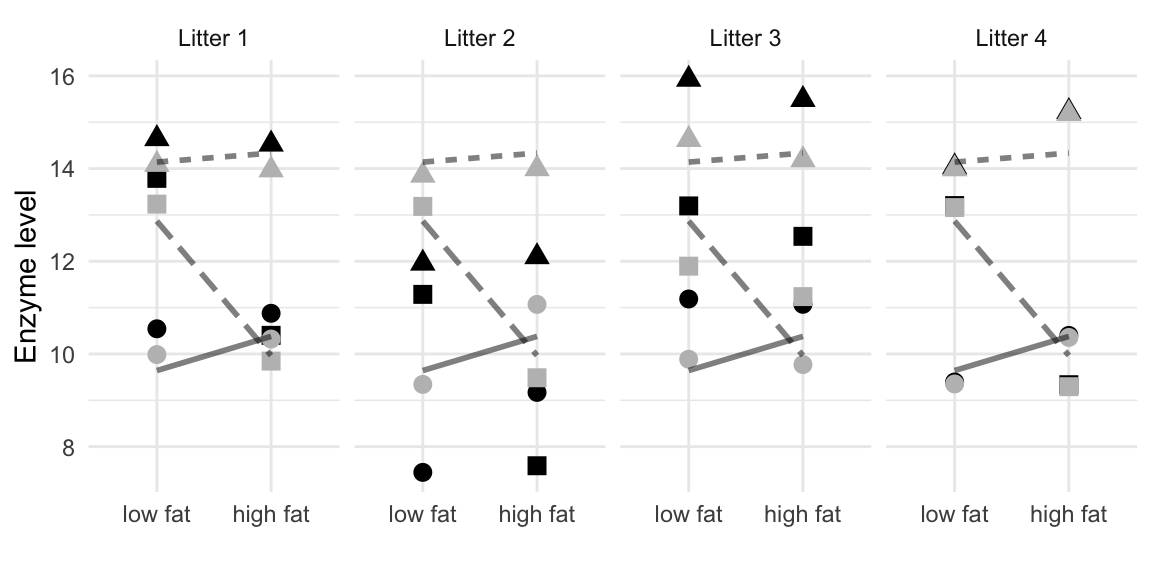
Figure 7.8: Enzyme levels for placebo (point), drug D1 (triangle), and drug D2 (square). Data are shown separately for each of four litters (blocks). Lines connect mean values over litters of same drug under low and high fat diets. Black points are raw data, grey points are enzyme levels adjusted for block effect.
ANOVA table
We analyse the resulting data using either analysis of variance with model specification y~drug*diet+Error(litter), or using a linear mixed model with specification y~drug*diet+(1|litter); both yield identical results, and the ANOVA table based on the linear mixed model is
| Sum Sq | Mean Sq | NumDF | DenDF | F value | Pr(>F) | |
|---|---|---|---|---|---|---|
| drug | 74.1 | 37.05 | 2 | 15 | 91.54 | 3.931e-09 |
| diet | 2.586 | 2.586 | 1 | 15 | 6.39 | 0.02319 |
| drug:diet | 15.41 | 7.707 | 2 | 15 | 19.04 | 7.646e-05 |
Blocking by litter reduces the residual variance from 1.19 for the non-blocked design in Chapter 6 to 0.64, the relative efficiency is RE(CRD,RCBD)= 1.87. Due to the increase in power, the Diet main effect is now significant.
Contrasts
The definition and analysis of linear contrasts presents no additional difficulties and the corresponding R function work exactly as for the two-way ANOVA in Section 6.5. For direct comparison with our previous results, we estimate the two interaction contrasts of Table 6.5 in the blocked design. They compare the difference in enzyme levels for D1 (resp. D2) under low and high fat diet to the corresponding difference in the placebo group; estimates and Bonferroni-corrected confidence intervals are shown in Table 7.2. As expected, the estimates are similar to those we found previously, but confidence intervals are substantially narrower due to the increase in precision.
| Contrast | Estimate | LCL | UCL |
|---|---|---|---|
| (D1 low - D1 high) - (Pl. low - Pl. high) | 0.54 | -1.04 | 2.13 |
| (D2 low - D2 high) - (Pl. low - Pl. high) | 3.64 | 2.06 | 5.22 |
7.3 Incomplete block designs
7.3.1 Introduction
The randomized complete block design can be too restrictive if the number of treatment levels is large or the available block sizes small. Fractional factorial designs offer a solution for factorial treatment structures by deliberately confounding some treatment effects with blocks (Chapter 9). For a single treatment factor, we can use incomplete block designs (IBD), where we deliberately relax the complete balance of the previous designs and use only a fraction of the treatments in each block.
The most important case of an IBD is the balanced incomplete block design (BIBD), where each pair of treatments is allocated to the same number of blocks, and so are then all individual treatments. This specific type of balance ensures that pair-wise contrasts are all estimated with the same precision, but precision decreases if more than two treatment groups are compared.
We illustrate this design with our drug example, where three drug treatments are allocated to 24 mice on a low fat diet. If the sample preparation is so elaborate that only two mice can be measured at the same time, then we have to run this experiment in twelve batches, with each batch containing one of three treatment pairs: (placebo, \(D1\)), (placebo, \(D2\)), or (\(D1\), \(D2\)). A possible balanced incomplete block design is shown in Figure 7.9, where each treatment pair is used on four batches, and the treatments are randomized independently to mice within each batch.The resulting data are shown in Table 7.3.
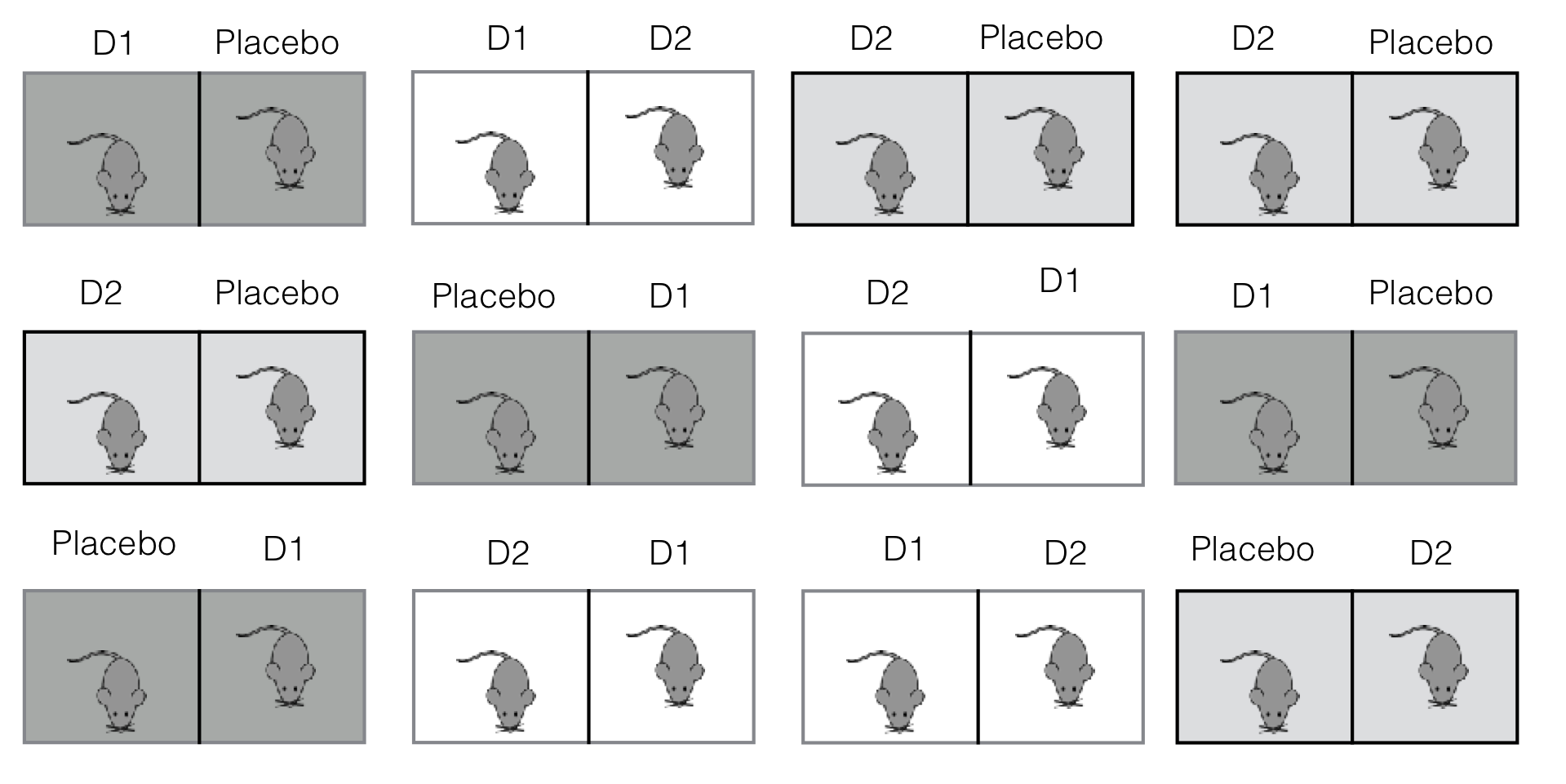
Figure 7.9: Experiment layout for a balanced incomplete block design with three drugs in twelve batches of size two.
|
|
|
7.3.2 Defining a BIBD
The key requirement of a BIBD is that all pairs of treatments occur the same number of times in the experiment. This requirement restricts the possible combinations of number of treatments, block size, and number of blocks. With three pairs of treatments as in our example, the number of blocks in a BIBD is restricted to be a multiple of three. The relations between the parameters of a BIBD are known as the two defining equations \[\begin{equation} rk = bs \quad\text{and}\quad \lambda\cdot (k-1) = r\cdot (s-1)\;. \tag{7.2} \end{equation}\] They relate the number of treatments \(k\), number of blocks \(b\), block size \(s\) to the number of times \(r\) that each treatment occurs and the number of times \(\lambda\) that each pair of treatments occurs in the same block. If these equations hold, then the corresponding design is a BIBD.
The equations are derived as follows: (i) with \(b\) blocks of \(s\) units each, the product \(bs\) is the total number of experimental units in the experiment. This number has to equal \(rk\), since each of the \(k\) treatments occurs in \(r\) blocks. (ii) moreover, each particular treatment occurs \(\lambda\) times with each of the remaining \(k-1\) treatments. It also occurs in \(r\) blocks, and is then combined in the block with \(s-1\) other treatment levels.
For our example, we have \(k=3\) treatment levels and each treatment occurs in \(r=8\) blocks, we have \(b=12\) blocks each of size \(s=2\), resulting in \(\lambda=4\) co-occurrences of each pair in the same block. This satisfies the defining equations and our design is a BIBD. In contrast, for only \(b=5\) batches, we are unable to satisfy the defining equations and no BIBD exists for this combination of number of blocks, block size, and number of treatments.
In order to generate a balanced incomplete block design, we need to find parameters \(s\), \(r\), and \(\lambda\) such that the defining equations (7.2) hold. A particularly simple way is by generating an unreduced or combinatorial design and use as many blocks as there are ways to select \(s\) out of \(k\) treatment levels. The number of blocks \(b\) is then \(b={k \choose s}=k!/(k-s)!s!\) and increases rapidly with \(k\).
We can often improve on the unreduced design, and generate a valid BIBD with substantially fewer blocks. Experimental design books sometimes contain tables of BIBDs for different number of treatment and block sizes, see for example (Cochran and Cox 1957). The simplest way for us is using R for finding a corresponding design, for example the function design.bib() of package agricolae. It requires a vector with the names of the treatment levels, and the desired block size \(s\); the option randomization=TRUE automatically produces a randomized experiment. For our example, we call bibd.design=design.bib(trt=c("Placebo", "D1", "D2"), k=2), and can extract the parameters as bibd.design$statistics and the treatment assignments as bibd.design$sketch.
As a further example, we consider constructing a BIBD for \(k=6\) treatments in blocks of size \(s=3\). The unreduced combinatorial design requires \(b={6 \choose 3}=20\) blocks. Not all of these are needed to fulfill the defining equations, however, and we find a much smaller design with \(b=10\) blocks using design.bib().
7.3.3 Analysis
Because not every treatment occurs in every block, the treatment and block main effects are no longer independent in an incomplete block design. This has three consequences for the practitioner: first, the treatment and block factor sums of squares are not independent, and while their sum remains the same, their individual values now depend on the order of the two factors in the model specification. Second, part of the information about treatment effects is now captured by the differences in blocks; in particular, there are two omnibus \(F\)-tests for the treatment factor, the first based on the inter-block information and located in the ANOVA table in the block-factor error stratum, and the second based—as for an RCBD—on the intra-block information and located in the residual error stratum. Third, the linear mixed model approach offers a more natural method for analysis that simultaneously uses the full information, and analysis of variance and mixed model results no longer concur exactly.
Linear model
We assume negligible block-by-treatment interaction, and the linear model for a balanced incomplete block design is the additive model \[ y_{ij} = \mu + \alpha_i + b_j + e_{ij}\;, \] where \(\mu\) is a general mean, \(\alpha_i\) the expected difference between the group mean for treatment \(i\) and the grand mean, \(b_j\sim N(0,\sigma_b^2)\) are the random block effects, and \(e_{ij}\sim N(0,\sigma_e^2)\) are the residuals. All random variables are mutually independent.
Not all block-treatment combinations occur in a BIBD, and some of the \(y_{ij}\) therefore do not exist. As a consequence, naive parameter estimators are biased, and more complex estimators are required for the parameters and treatment group means. We forego a more detailed discussion of these problems and rely on statistical software like R to provide appropriate estimates for BIBDs.
Intra- and inter-block information
In an RCBD, we can estimate any treatment contrast and all effects independently within each block, and then average over blocks. We can use the same intra-block analysis for a BIBD by estimating contrasts and effects based on those blocks that contain sufficient information and averaging over these blocks. The resulting estimates are free of block effects.
In addition, the block totals—calculated by adding up all response values in a block—also contain information about contrasts and effects if the block factor is random. This information can be extracted by an inter-block analysis. We again refrain from discussing the technical details, but instead provide some intuition for the recovery of inter-block information.
We consider our example with three drugs in blocks of pairs and are interested in estimating the average enzyme level under the placebo treatment. Using the intra-block information, we would adjust each response value by the block effect, and average over the resulting adjusted placebo responses. For the inter-block analysis, first note that the true treatment group means are \(\mu+\alpha_1\), \(\mu+\alpha_2\), and \(\mu+\alpha_3\) for placebo, \(D1\), and \(D2\), respectively. The block totals for the three types of block are then (placebo, \(D1\))\(=T_{P,D1}=\mu+\alpha_1+\mu+\alpha_2\), (placebo, \(D2\))\(=T_{P,D2}=\mu+\alpha_1+\mu+\alpha_3\), and (\(D1\), \(D2\))\(=T_{D1,D2}=\mu+\alpha_2+\mu+\alpha_3\), respectively and each type of block occurs the same number of times.
The treatment group mean for placebo can then be calculated from the block totals as \((T_{P,D1}+T_{P,D2}-T_{D1,D2})/2\), since \[\begin{align*} \frac{T_{P,D1}+T_{P,D2}-T_{D1,D2}}{2} &= \frac{2\mu+\alpha_1+\alpha_2}{2} + \frac{2\mu+\alpha_1+\alpha_3}{2} - \frac{2\mu+\alpha_2+\alpha_3}{2} \\ &=\left(4\mu+2\alpha_1+\alpha_2+\alpha_3\right) - \left(2\mu+\alpha_2+\alpha_3\right) = \mu+\alpha_1\;. \end{align*}\] In our illustration (Fig. 7.9), this corresponds to taking the sum over responses in the light and dark grey blocks, and subtracting the responses in the white blocks. Note that no information about treatment effects is contained in the block totals of an RCBD.
Analysis of variance
Based on the linear model, the specification for the analysis of variance is y~drug+Error(block) for a random block factor, the same as for an RCBD. The resulting ANOVA table has again two error strata, and the block error stratum contains the inter-block information about the treatment effects. With two sums of squares, the analysis provides two different omnibus \(F\)-tests for the treatment factor:
##
## Error: block
## Df Sum Sq Mean Sq F value Pr(>F)
## drug 2 14.82 7.409 3.415 0.0788 .
## Residuals 9 19.53 2.170
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Error: Within
## Df Sum Sq Mean Sq F value Pr(>F)
## drug 2 56.30 28.151 98.06 2.69e-07 ***
## Residuals 10 2.87 0.287
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The first \(F\)-test is based on the inter-block information about the treatment, and is in general (much) less powerful than the second \(F\)-test based on the intra-block information. Both \(F\)-tests can be combined to an overall \(F\)-test taking account of all information, but the inter-block \(F\)-test is typically ignored in practice and only the intra-block \(F\)-test is used and reported; this approximation leads to a (often small) loss in power.
The dependence of the block- and treatment factors must be considered for fixed block effects. The correct model specification contains the blocking factor before the treatment factor in the formulae, and is y~block+drug for our example. This model yields an analysis identical to the intra-block analysis for random block factors. The model y~drug+block, on the other hand, yields an entirely different ANOVA table and an incorrect \(F\)-test. We provide more details of these problems of fixed-effects analysis of incomplete data in Chapter ??.
Mixed model analysis
With the advent of powerful statistical software, we can avoid many of the limitations of the analysis of variance by using a linear mixed model approach for our analysis. It does not require error strata to cope with the two variance components, and provides a single estimate of the drug effects and treatment group means based all available information. The resulting degrees of freedom are no longer integers and resulting \(F\)-tests and \(p\)-values can deviate from the classical analysis of variance. We specify the model as y~drug+(1|block); this results in the ANOVA table
| Sum Sq | Mean Sq | NumDF | DenDF | F value | Pr(>F) | |
|---|---|---|---|---|---|---|
| drug | 55.64 | 27.82 | 2 | 10.81 | 96.57 | 1.277e-07 |
We note that the sum of squares and the mean square estimates are slightly larger than for the aov() analysis, because the between-blocks information is taken into account. This also results in a slightly larger value of the \(F\)-statistic. For more complex designs, denominator degrees of freedom for an \(F\)-test cannot be calculated exactly, and several approximations can be used. The conservative Kenward-Roger approximation yields the same degrees of freedom as the standard ANOVA approach for balanced designs and we use it throughout.
This is our first example of a design in which the deliberate violation of complete balance still allows an analysis of variance, but where a mixed model analysis gives advantages both in the calculation but also the interpretation of the results.
7.3.4 Contrasts
Using R, the estimation of treatment contrasts proceeds exactly as in our previous designs. Using either aov() or lmer() for estimating the linear model, we estimate the corresponding marginal means by emmeans() and define contrasts as before. The marginal means from aov() are based exclusively on the intra-block information and can differ from those based on lmer(). This also affects the contrast estimates and their confidence intervals. For our example, we calculate three contrasts comparing each treatment respectively their average to placebo. The results are shown in Table 7.4.
| Contrast | Estimate | SE | df | LCL | UCL |
|---|---|---|---|---|---|
| ANOVA: aov(y~drug+Error(block)) | |||||
| D1 - Placebo | 4.28 | 0.31 | 10.00 | 3.59 | 4.97 |
| D2 - Placebo | 2.70 | 0.31 | 10.00 | 2.01 | 3.39 |
| (D1+D2)/2 - Placebo | 3.49 | 0.27 | 10.00 | 2.90 | 4.09 |
| Mixed model: lmer(y~drug+(1|block)) | |||||
| D1 - Placebo | 4.22 | 0.31 | 10.81 | 3.54 | 4.90 |
| D2 - Placebo | 2.74 | 0.31 | 10.81 | 2.06 | 3.42 |
| (D1+D2)/2 - Placebo | 3.48 | 0.27 | 10.81 | 2.89 | 4.07 |
Estimation of all treatment contrasts is unbiased and contrast variances are free of the block variance in a BIBD. The variance of a contrast estimate is \[ \text{Var}(\hat{\Psi}(\mathbf{w})) = \frac{s}{\lambda k}\sigma_e^2\sum_{i=1}^kw_i^2\;. \] where each treatment is assigned to \(r\) units. The corresponding contrast in an RCBD with the same number \(n=r\) of replicates for each treatment (but lower number of blocks) has variance \(\sigma_e^2\sum_i w_i^2/n\) and we can interpret the term \[ \frac{\lambda k}{s}=r\frac{k}{s}\frac{s-1}{k-1}=n^* \] as the “effective sample size” in each treatment group for a BIBD. This effective sample size is smaller than the actual replication \(r\) since some information is lost because we cannot fully eliminate the block variances from the group mean estimates.
The relative efficiency of a BIBD compared to an RCBD with the same number of units per treatment is \[ \text{RE}_{\Psi}(\text{BIBD},\text{RCBD}) = \frac{r\frac{k}{s}\frac{s-1}{k-1}}{r} = \frac{n^*}{r}\;, \] which is \((3-1)/(2-1)\cdot 2/3=4/3\) for our example, and our BIBD would require about 33% more samples to achieve the same precision as an RCBD. The fewer treatments fit into a single block, the lower the relative efficiency.
7.3.5 Power analysis and sample size
We conduct sample size determination and power analysis for a balanced incomplete block design in much the same fashion as for a randomized complete block design. The denominator degrees of freedom for the omnibus \(F\)-test are \(\text{df}_\text{res}=bk-b-k+1\). The noncentrality parameter is \[ \lambda = r\cdot k \cdot \frac{s-1}{k-1}\frac{k}{s}\cdot f^2=k\cdot n^*\cdot f^2\;, \] which is again of the form “total sample size times effect size”, but the total sample size uses the effective sample size per group to adjust for the incompleteness. For the same number \(r\) of units per treatment, the noncentrality parameter of a BIBD is thus smaller than the parameter for an RCBD, and the BIBD has lower power.
7.3.6 Reference designs
The reference design or design for differential precision (Cox 1958, 238) is a variation of the BIBD that is useful when a main objective is comparing treatments to a control (or reference) group. One concrete application is a microarray study, where several conditions are to be tested, but only two dyes are available for each microarray. If all pairwise contrasts between treatment groups are of equal interest, then a BIBD shown in Figure 7.10(A, right) is a reasonable option. If all treatments are to be compared to a control condition, a suitable reference design is given in Figure 7.10(A, left).
Another application of reference designs is the screening of several new treatments against a standard treatment. In this case, selected treatments might be compared among each other in a subsequent experiment, and removal of unpromising candidates in the first round might reduce the later efforts.
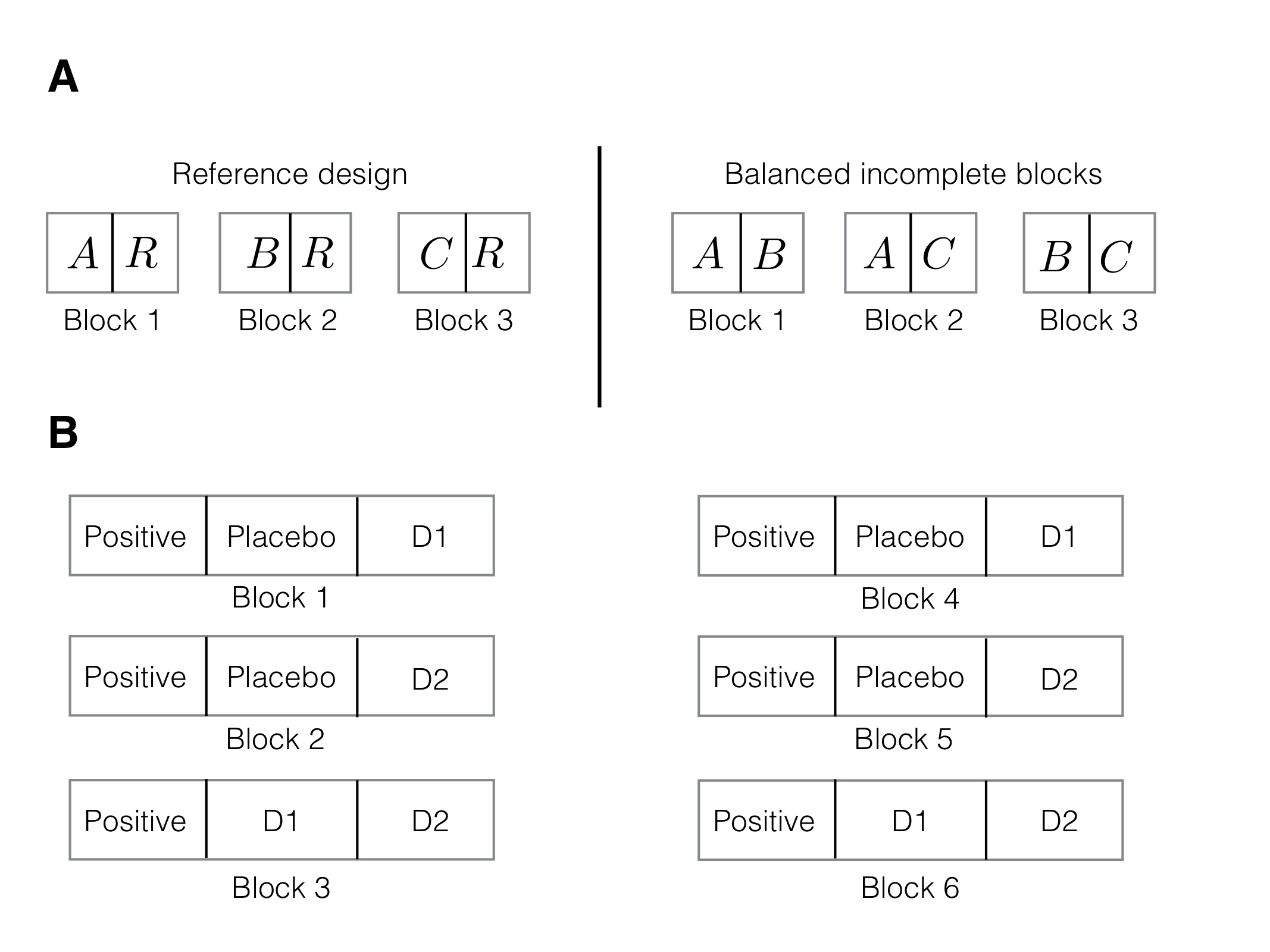
Figure 7.10: A: Reference design for three treatments A,B,C and a reference condition R (left) and BIBD without reference condition (right). B: reference design for three treatments and common positive control as reference condition with block size three.
Generating a simple reference design is straightforward: since each block of size \(s\) contains the reference condition, it has \(s-1\) remaining experimental units for accommodating a subset of the \(k-1\) non-control treatments. This translates into finding a BIBD for \(k-1\) treatments in blocks of size \(s-1\). The main analysis of this design is based on Dunnett-type contrasts, where each treatment level is compared to the reference level. The same considerations as for a CRD with Dunnett-type contrasts then apply, and we might think about using more than one slot per block for the reference condition to improve precision. A balance has then to be struck between improving the comparisons within each block by using more than one replicate of the control condition, and the need to use more blocks and introduce more uncertainty by having smaller block sizes available for the treatment conditions.
For our drug example, we might introduce a positive control condition in addition to the placebo group. A reference design with block size \(s=3\) then requires \(b=3\) blocks, with treatment assignments (positive, placebo, \(D1\)) for the first, (positive, placebo, \(D2\)) for the second, and (positive, \(D1\), \(D2\)) for the third block; this is simply a BIBD with \(s=2\) and \(k=3\), augmented by adding the positive treatment to each block. A two-fold replication is shown in Figure 7.10 (B).
The same principle extends to constructing reference designs with more than one reference level. In our example, we might want to compare each treatment to positive and placebo (negative) control groups, and we accommodate the remaining treatment factor levels in blocks of size \(k-2\). If the available block size is \(k=3\), then no remaining treatment levels co-occur in the same block. Comparisons of these levels to reference levels than profit from blocking, but comparisons between the remaining treatment levels do not.
7.4 Multiple blocking factors
7.4.1 Introduction
The unit structure of (G)RCBD and BIBD experimental designs consists of only two unit factors: the blocks and the nested experimental units. We now discuss unit structures with more than one blocking factor; these can be nested or crossed.
7.4.2 Nesting blocks
Nesting blocking factors allows us to replicate already blocked designs, to combine several properties for grouping, and to disentangle the variance components of different levels of grouping. Nesting is sometimes also dictated by the logistics of the experiment, as exemplified by the example in this section. We consider only designs with two nested blocking factors, but our discussion directly translates to longer chains of nested factors.
We saw that we can replicate a completely randomized design by crossing its treatment structure with a factor to represent the replication; in our example, we used a CRD with three drugs in two laboratories, each laboratory providing one replicate of the CRD. The same idea allows replicating more complex designs: to replicate our RCBD for drug comparisons blocked by litter in two laboratories, for example, we again cross the treatment structure with a new factor (Lab) with two levels. Since each litter is unique to one laboratory, (Litter) from the original RCBD is nested in (Lab), and both are crossed with Drug, since each drug is assigned to each litter in both laboratories. This yields the treatment and design structures shown in Figure 7.11(left,center-left). The experiment design in Figure 7.11(center-right) then contains two block-by-treatment interactions, and the three-way interaction is an interaction of a lab-specific litter with the treatment factor. This experiment cannot by analysed using analysis of variance, because we cannot specify the block-by-treatment interactions; the linear mixed model is y~drug+(1|lab)+(1|lab:drug)+(1|lab:litter)+(1|lab:litter:drug).
If we careful select the blocking factors, we can assume these interactions to be negligible and arrive at the much simpler experiment design in Figure 7.11(right). Our experiment again uses 24 mice, with four litters per laboratory, and each litter a block with one replicate per treatment. The model specification is y~drug+Error(lab/litter) for an analysis of variance, and y~drug+(1|lab/litter) (or equivalently y~drug+(1|lab)+(1|lab:litter)) for a linear mixed model. Since more than one litter is used per lab, the linear mixed model directly provides us with estimates of the between-lab the between-to-litter (within lab) variance components. This provides valuable insight into the sources of variation in this experiment and would allow us, for example, to conclude if the blocking by litter was successful on its own, and how discrepant values for the same drug are between different laboratories. Since the two blocks are nested, omnibus \(F\)-test and contrasts for Drug are estimated within each litter and then averaged over litters within labs.
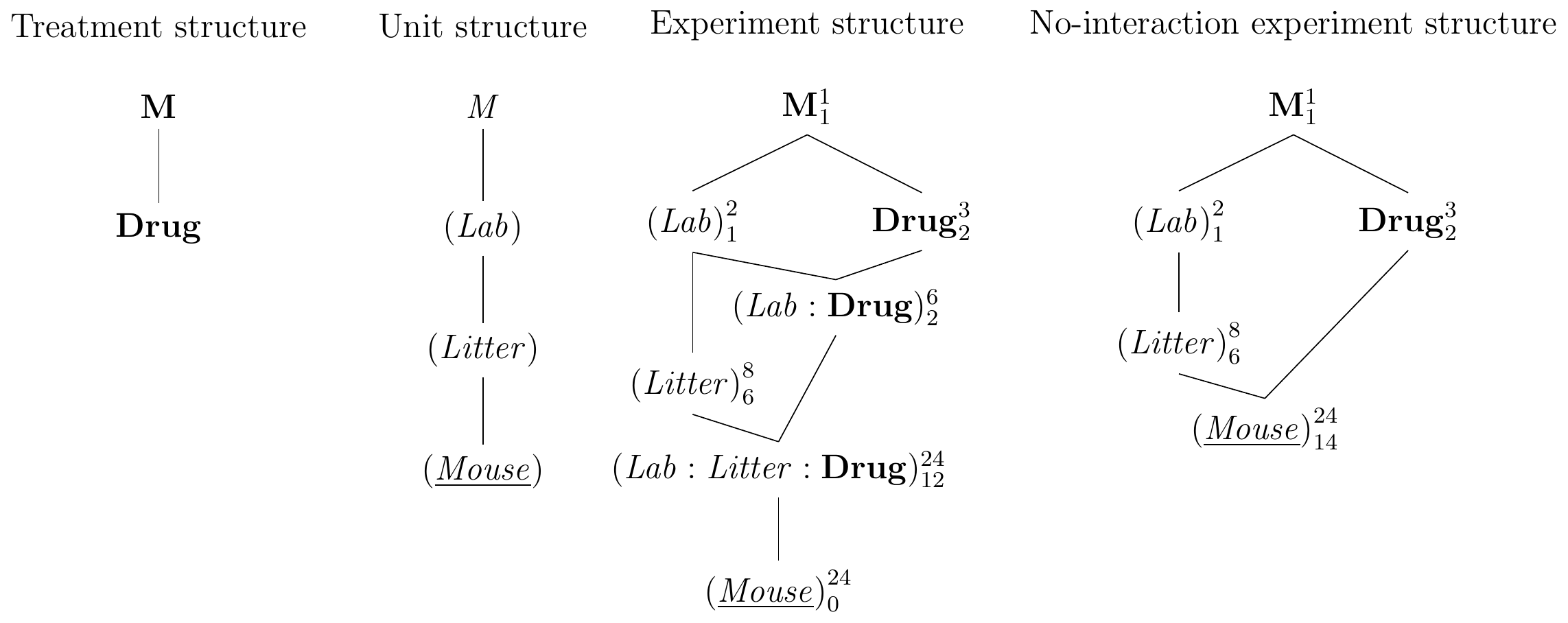
Figure 7.11: Randomized complete block design for determining effect of two different drugs and placebo treatments on enzyme levels in mice, replicated in two laboratories. Left: treatment structure. Center-left: unit structure with three nested factors. Center-right: full experiment structure with block-by-treatment interactions. Right: experiment structure if interactions are negligible.
This type of design can be extended to an arbitrary number of nested blocks and we might use two labs, two cages per lab, and two litter per cage for our example. As long as each nested factor is replicated, we are able to estimate corresponding variance components. If a factor is not replicated (e.g., we use a single litter per lab), then there are no degrees of freedom for the nested blocking factor, and the effects of both blocking factors are completely confounded. Effectively, such a design uses a single blocking factor, where each level is a combination of lab and litter.
We should keep in mind that variance components are harder to estimate than averages, and estimates based on low replication are very imprecise. This is not a problem if our goal is removal of variation from treatment contrasts, but sufficient replication is necessary if estimation of variance components is a primary goal of the experiment. Sample size determination for variance component estimation is covered in more specialized texts.
7.4.3 Latin squares
Nesting blocking factors essentially results in replication of (parts of) the design. In contrast, crossing blocking factors allows us to control several sources of variation simultaneously. The most prominent example is the latin square design (LSD), which consists of two crossed blocking factors simultaneously crossed with a treatment factor, such that each treatment level occurs once in each level of the two blocking factors.
For example, we might be concerned about the effect of litters on our drug comparisons, but suspect that the position of the cage in the rack also affects the observations. The latin square design removes both between-litter and between-cage variation from the drug comparisons. For three drugs, this design requires three litters of three mice each, and three cages. Crossing litters and cages results in one mouse per litter in each cage. The drugs are then randomized on the intersection of litters and cages (i.e., mice) such that each drug occurs once in each cage and once in each litter. An example layout of this design is shown in Figure 7.12A, where the two blocking factors are shown as rows and columns. Each treatment occurs exactly once per row and once per column and the latin square design imposes two simultaneous constraints on the randomization of drug on mice.
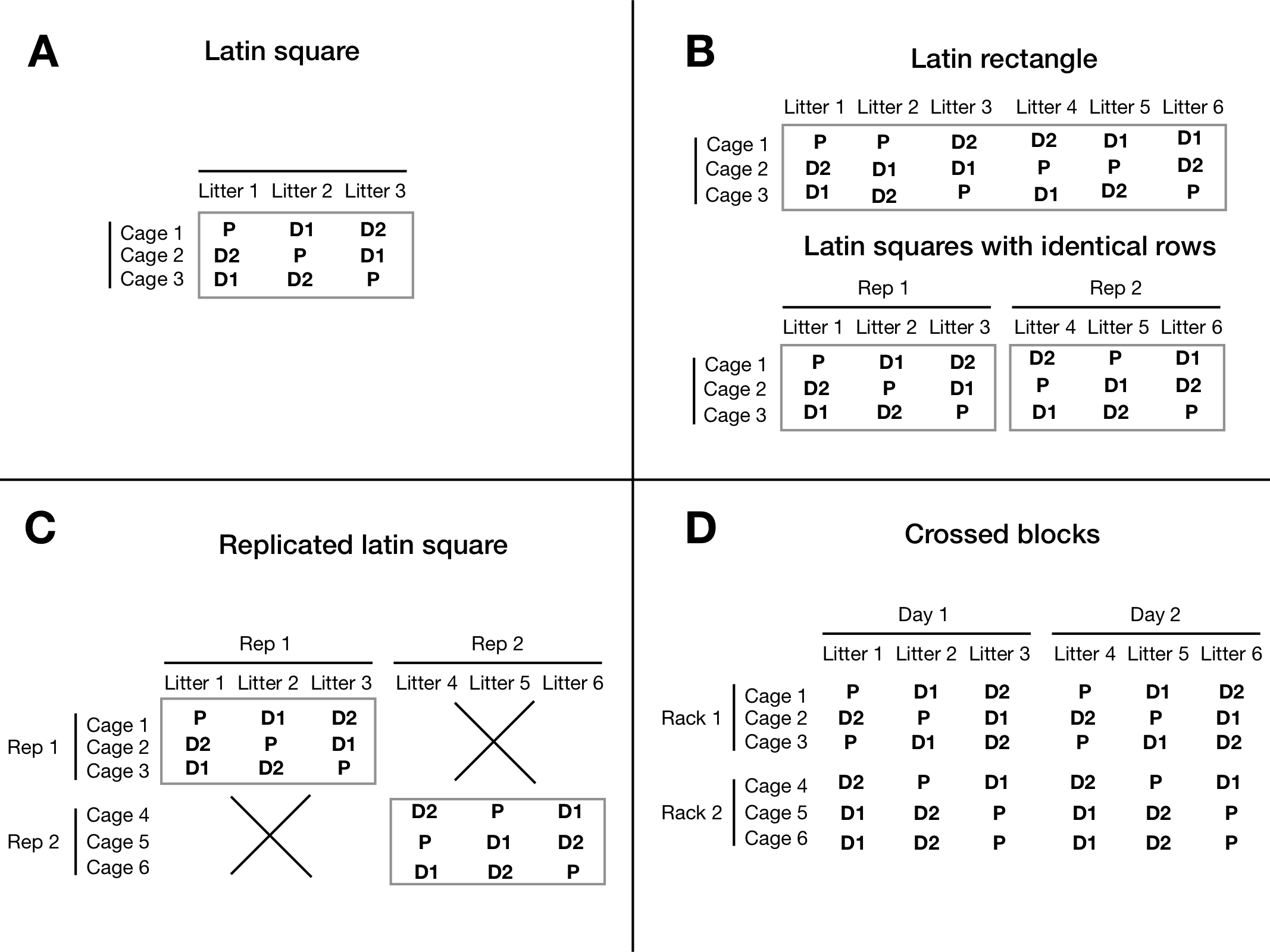
Figure 7.12: A: Experiment layout for latin square design using cage and litter as two crossed block. Three drug treatment levels are randomly assigned to cells such that each level occurs exactly once per cage and litter. B: full replication with new rows and new columns. C: possible replications keeping the same cages, and using more litters. D: two fully crossed blocks.
The Hasse diagrams for this design are shown in Figure 7.13. The crucial observation is that none of the potential block-by-block or block-by-treatment interactions can be estimated, since each combination of block levels or block-treatment combinations occurs only once. In particular, the intersection of a cage and a litter is a single mouse, and thus (Cage:Litter) is completely confounded with (Mouse) for our example.
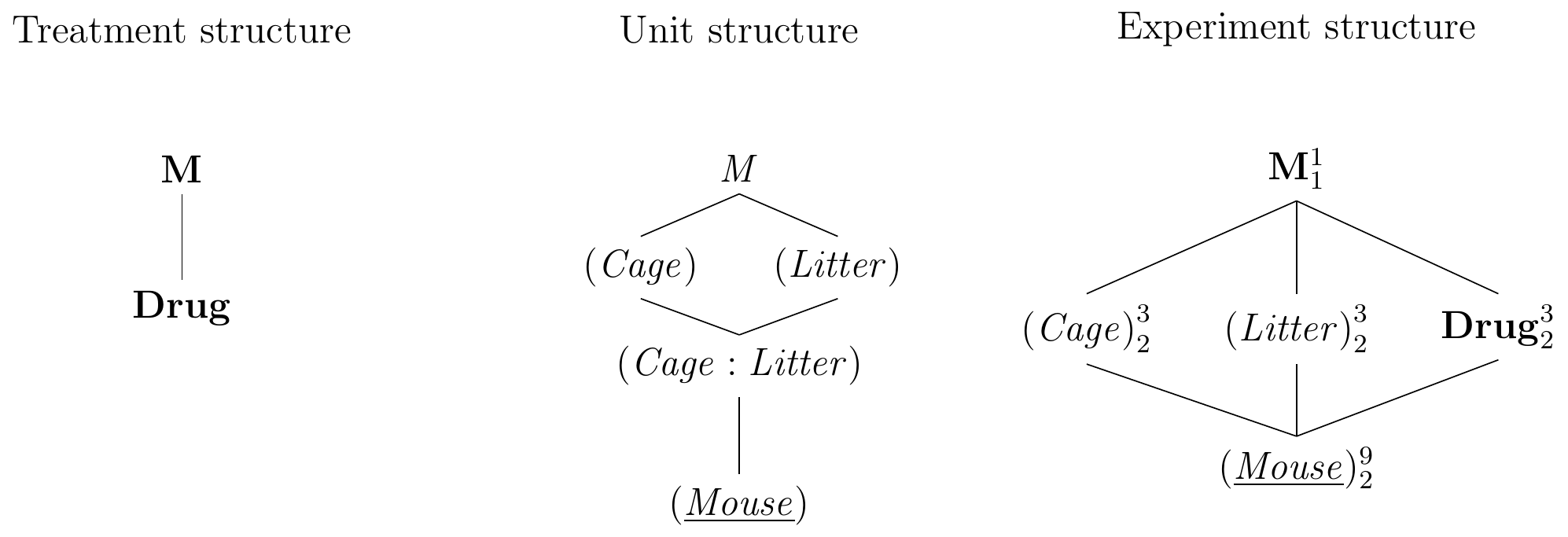
Figure 7.13: Latin square with cage and litter as random row/column effects and three drug treatment levels.
With all interactions negligible, data from a latin square design are analysed by an additive model
\[
y_{ijk} = \mu + \alpha_i + c_j + r_k + e_{ijk}\;,
\]
where \(\mu\) is a general mean, \(\alpha_i\) the treatment parameters, and \(c_j\) (resp. \(r_k\)) are the random column (resp. row) parameters. This model is again in direct correspondence to the experiment diagram and is specified as y~drug+Error(cage+litter), respectively y~drug+(1|cage)+(1|litter).
We can interpret the latin square design as a blocked RCBD: ignoring the cage effect, for example, the experiment structure is identical to an RCBD blocked by litter. Conversely, ignoring the litter effect, we have an RCBD blocked on cages. The remaining blocking factor now blocks the whole RCBD. The requirement of having each treatment level once per cage and litter requires that the number of cages must equal the number of litters, and that both must also equal the number of treatments. These constraints pose some complications for the randomization of treatment levels to mice, and randomization is usually done by software such as the agricolae and dae packages in R; many older books on experimental design also contain tables latin square designs with few treatment levels.
7.4.3.1 Replicating latin squares
A design with a single latin square often suffers from insufficient replication: for \(k=2\), \(k=3\), and \(k=4\) treatment levels, we only have \(0\), \(2\), and \(6\), residual degrees of freedom, respectively. We demonstrate several options for generating \(r\) replicates of a latin square design. In each case, the Hasse diagram allows us to quickly calculate the resulting error degrees of freedom and to compare them between the designs. Moreover, the specification of an appropriate linear model is again quickly derived from the diagrams.
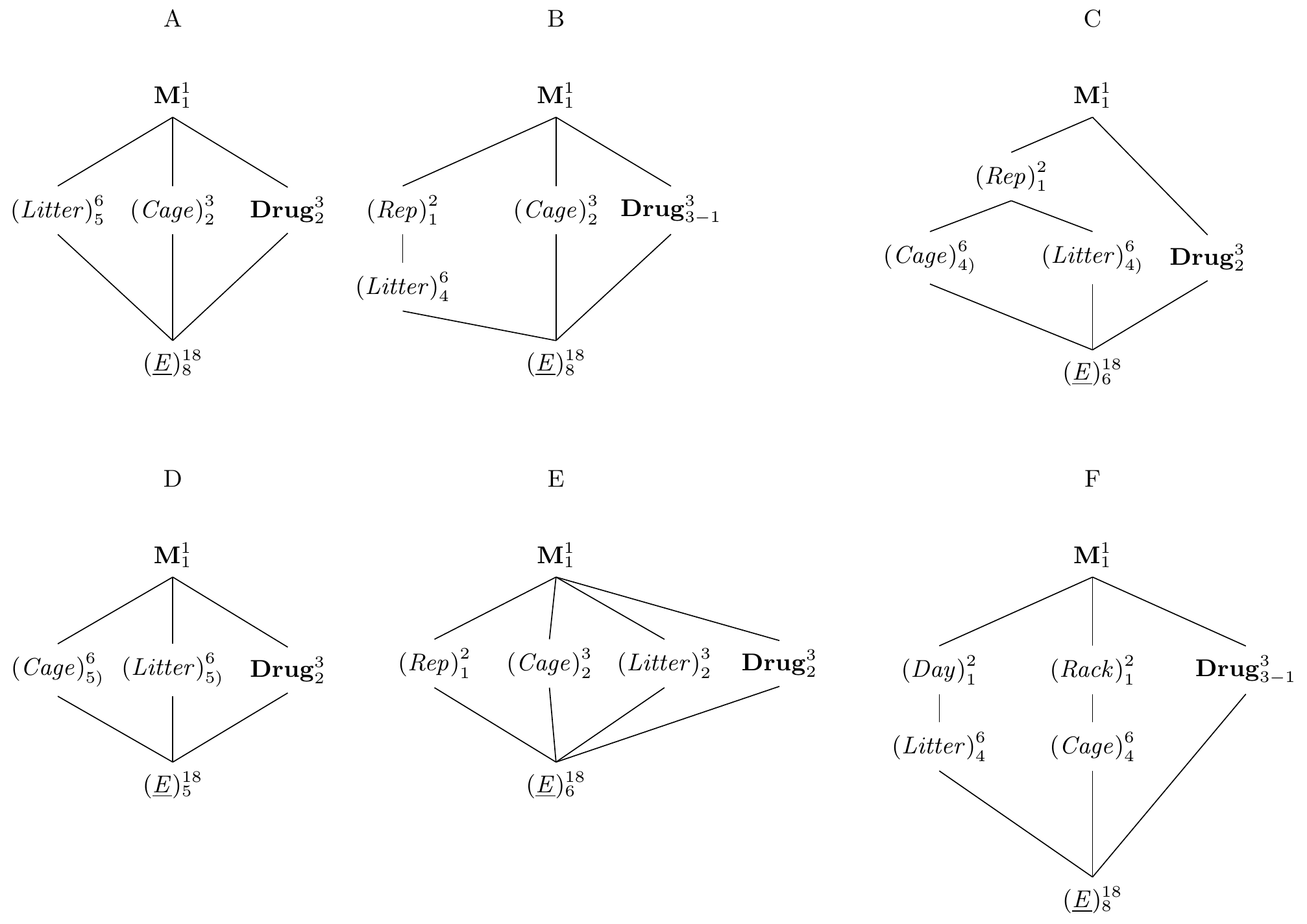
Figure 7.14: Replication of latin square design. A: latin rectangle with six columns and three rows. B: two sets of three columns with identical rows. C: new rows and columns in each replicate. C: two independent latin squares with identical treatments. D: fully crossed blocks, not a latin square. E: keeping both row and column levels with two independently randomized replicates. F: independent replication of rows and columns.
First, we might consider replicating columns while keeping the rows, using \(rk\) column factor levels instead of \(k\). The same logic applies to keeping columns and replicating rows, of course. The two experiments in Figure 7.12B illustrate this design for a two-fold replication of the \(3\times 3\) latin square, where we use six litters instead of three, but keep using the same three cages in both replicates. In the top part of the panel, we do not impose any new restrictions on the allocation of drugs and only require that each drug occurs the same time in each cage, and that each drug is used with each litter. In particular, the first three columns do not form a latin square in themselves. This design is called a , and its experiment structure is shown in Figure 7.14A with model specification y~drug+Error(cage+litter) or y~drug+(1|cage)+(1|litter).
We can also insist that each replicate forms a proper latin square in itself. That means we group the columns in two groups of three as shown in the bottom of Figure 7.12B. In the diagram in Figure 7.14B, this is reflected by a new grouping factor (Rep) with two levels, in which the column factor (Litter) is nested. The model is specified as y~drug+Error(cage+rep/litter) or y~drug+(1|cage)+(1|rep/litter).
By nesting both blocking factors in the replication, we find yet another useful design that uses different row and column factor levels in each replicate of the latin square as shown in Figure 7.12C. The corresponding diagram is 7.14C and yields the model y~drug+Error(rep/(cage+litter)) or y~drug+(1|rep)+(1|rep/cage)+(1|rep/litter).
We can also replicate both rows and columns (potentially with different numbers of replication) without resctricting any subset of units to a latin square. The experiment is shown in Figure 7.12D and extends the latin rectangle to rows and columns. Here, none of the replicates alone forms a latin square, but all rows and columns have the same number of units for each treatment level. The diagram is shown in Figure 7.14C with model specification y~drug+Error(cage+litter) or y~drug+(1|cage)+(1|litter). Using this design but insisting on a latin square in each of the four replicates yields the diagram in Figure 7.12F with model y~drug+Error(rack/cage+day/litter) or y~drug+(1|rack/cage)+(1|day/litter)
Finally, we might also use the same row and column factor levels in each replicate. This is rarely the case for random block factors, but we could use our three drugs with three measurement devices and three laboratory technicians, for example, such that each technician measures one mouse for each treatment in each of the three devices. Essentially, this fully crosses the latin square with a new blocking factor and leads to the model specification y~drug+Error(rep+device+tech) or y~drug+(1|rep)+(1|device)+(1|tech).
7.4.4 Power analysis and sample size
The determination of sample size and power for a crossed-block design requires no new ideas. The noncentrality parameter \(\lambda\) is again the total sample size times the effect size \(f^2\), and we find the correct degrees of freedom for the residuals from the experiment diagram.
For a single latin square, the sample size is necessarily \(k^2\) and increases to \(r\cdot k^2\) is we use \(r\) replicates. The residual degrees of freedom then depend on the strategy for replication. The residual degrees of freedom and the noncentrality parameter are shown in Table 7.5 for a single latin square and three replication strategies: using the same levels for row and column factors, using the same levels for either row or column factor and new levels for the other, and using new levels for both blocking factors. The numerator degrees of freedom are \(k-1\) in each case.
| Design | NCP \(\lambda\) | \(\text{df}_{\text{res}}\) |
|---|---|---|
| Single latin square | \(\lambda=k^2\cdot f^2\) | \((k-1)(k-2)\) |
| LS, same rows, same columns | \(\lambda=r\cdot k^2\cdot f^2\) | \((k-1)(r(k-1)-3)\) |
| LS, same rows, new columns | \(\lambda=r\cdot k^2\cdot f^2\) | \((k-1)(rk-2)\) |
| LS, new rows, new columns | \(\lambda=r\cdot k^2\cdot f^2\) | \((k-1)(r(k-1)-1)\) |
7.4.5 Balanced incomplete designs with two block factors: Youden designs
The latin square design requires identical number of levels for both row and column factor. We can two blocking factors with a balanced incomplete block design to reduce the required number of levels for one of the two blocking factors. These designs are called Youden squares and only use a fraction of the treatment levels in each column (resp. row) and the full set of treatments in each row (resp. column).
The idea was first proposed by Youden in 1937 for studying inoculation of tobacco plants against the mosaic virus (Youden 1937). In the experiment, seven plants were considered, and seven different inoculation treatments. It was suspected that the height of a plant’s leaf influences the effect of the virus, and leaf height was used as a second blocking factor with three levels (low, middle, high). Crossing the two blocking factors leads to a \(3\times 7\) layout. The treatment levels are randomly allocated such that each treatment occurs once per leaf height (i.e., each inoculation occurs once in each column), and the columns form a balanced incomplete block design of seven treatments in seven blocks of size three. A possible experiment layout is shown in Figure 7.15.
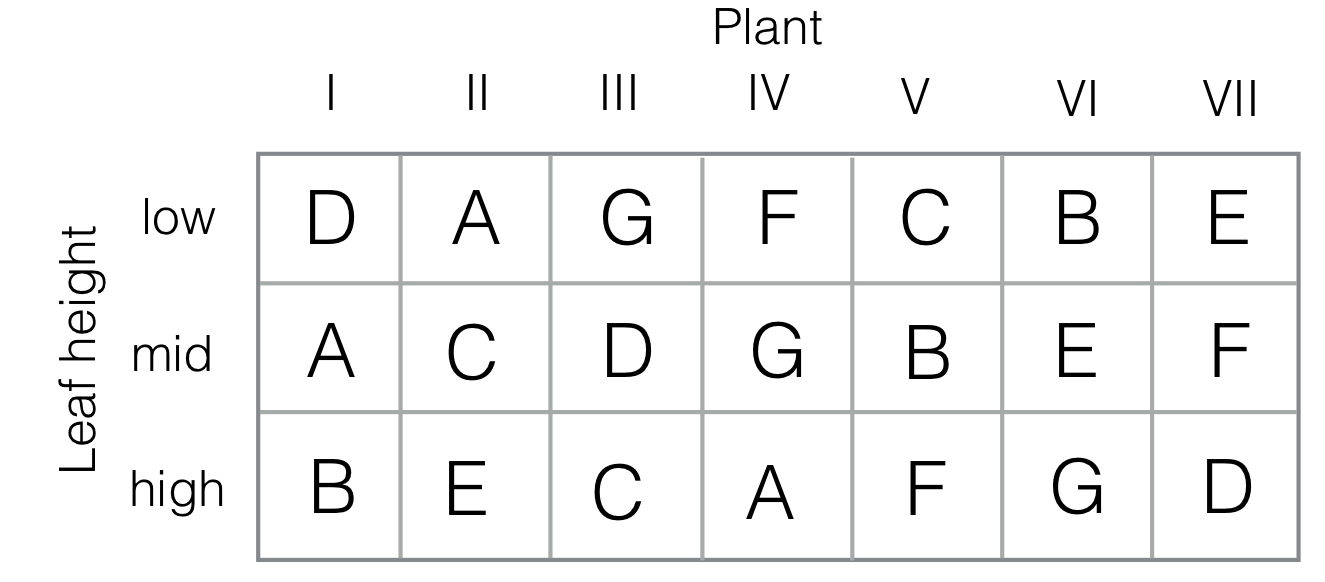
Figure 7.15: Experiment layout for Youden square. Seven plants are considered (I-VII), with seven different inoculation treatments (A-G). In each plant, three leafs at different heights (low/mid/high) are inoculated. Each inoculation occurs once at each height, and the assignment of treatments to plants forms a balanced incomplete block design.
For constructing a Youden square, we can augment the smaller row factor with dummy levels to match the number of levels of the column and the treatment factor. For our example, this means augmenting the leave height by four additional levels. We then construct a latin square, and delete from it the four augmented levels. The design in Figure 7.15 was constructed using the design.youden() function of package agricolae in R as design.youden(trt=c("A","B","C","D","E","F","G"), r=3).
References
Cochran, W. G., and G. M. Cox. 1957. Experimental Designs. John Wiley & Sons, Inc.
Cox, D R. 1958. Planning of Experiments. Wiley-Blackwell.
Fisher, R. 1971. The Design of Experiments. 8th ed. Hafner Publishing Company, New York.
Karp, Natasha A. 2018. “Reproducible preclinical research—Is embracing variability the answer?” PLOS Biology 16 (3): e2005413. https://doi.org/10.1371/journal.pbio.2005413.
Perrin, S. 2014. “Make mouse studies work.” Nature 507: 423–25.
Samuels, Myra L, George Casella, and George P McCabe. 1991. “Interpreting Blocks and Random Factors.” Journal of the American Statistical Association 86 (415). American Statistical Association: 798–808. http://www.jstor.org/stable/2290415.
Watt, L. J. 1934. “Frequency Distribution of Litter Size in Mice.” Journal of Mammalogy 15 (3): 185–89. https://doi.org/10.2307/1373847.
Youden, W. J. 1937. “Use of incomplete block replications in estimating tobacco-mosaic virus.” Cont. Boyce Thompson Inst. 9: 41–48.